Bioinformatics Tutorials & Lessons: Use SWISSPROT to search for a specific protein:
ADVERTISEMENTS
Bioinformatics Tutorials & Lessons:Tutorials & Lessons: use SWISSPROT to search for a specific protein:
Let say that you have a specific protein and you want to do some research about it including:
1- Informations about organisms that have this protein.
2- The function of this protein.
3- The protein sequence.
4- Complete references about this protein...............etc
And a lot of athor features and informations.
That protein is for example "Myosin", you can choose any protein you're interested in.
The first step to do is to enter to the SWISSPROT website at expasy:
1- Enter the site directly from here or go to google and write SWISSPROT, the first website at expasy is the SWISSPROT website, you'll see something like this:
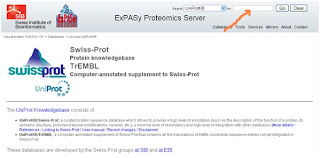
2- Enter your protein name in the search box shown by the red arrow in the picture above.
* In this tutorial i'm searching for the protein "myosin" for example.
3- Click the GO button to start the search.
4- You will see the result page like bellow:
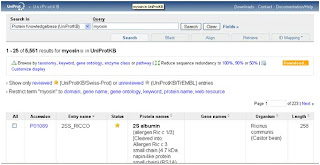
The information provided by this result is to huge and not accurate like related proteines, so we need to set a couple of things to get the results we need.
5- To do that you need to click on fields shown bellow:
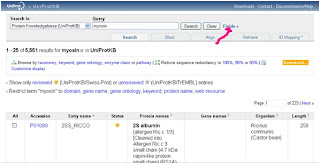
You will see this:
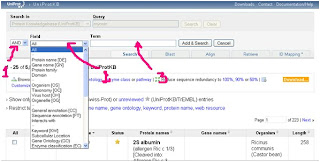
From 1 shown by the arrow you can choose AND, NOT, OR
AND: means that you will add something like organism name for example.
NOT: means that you can eliminate searches that contain the word you'll write, like eliminate an organism.
OR: will searches for example for myosin OR actin if you want to.
From 2 we can choose our field like protein name, organism, gene name...etc
From 3 the term section, we can add the word we need to add it to search.
6- We will set AND and Protein name from the field dropdown menu.
7- In the term section we write Myosin to search only for Protein wich names are Myosin and exclude related proteines.
8- We click Add & Search, and we will see this:
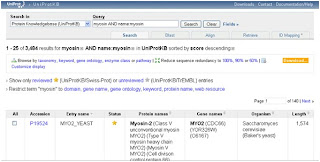
We remark that the number of hits had dropped down and also this hits shows only proteines with protein name containing the word Myosin.
Let's say for example that you want only the protein in a specific organism like Homo Sapiens, then we will repeat the steps from 5 to 8 by clicking "field", choosing AND, Organism from the field dropdown menu and write Homo Sapiens in the Term field.
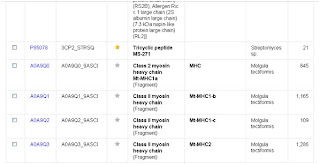
9- we click Add & Search and we'll get this:
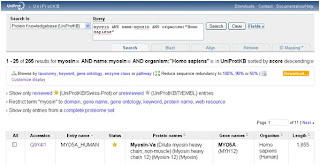
As you can see that the numbet of hits has dropped from more than 200 in the first search to 11 here.
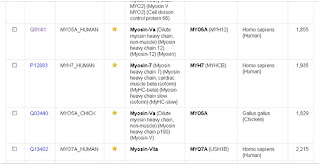
Because the protein Myosin have several chains, we will choose for example Myosin-Va.
10- By clicking the accession number shown bellow you will be taken to the information of this entry.

The informations about this protein are classed by category:
* Names and origins.
* Protein attributes.
* General anotations (protein function, subunits structure...etc).
* Refferences.
Our interest for now is the Sequences section, where we'll find the protein sequence.
We can see the sequence bellow:
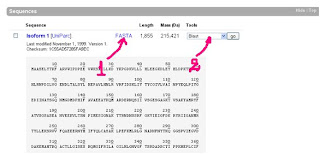
To see the protein sequence without numbers within lines click on 1 shown by the red arrow
To do a blast search for similarities with other proteines choose Blast and click go like shown 2 by the red arrow.
Any comment or question, you're welcome.
1- Informations about organisms that have this protein.
2- The function of this protein.
3- The protein sequence.
4- Complete references about this protein...............etc
And a lot of athor features and informations.
That protein is for example "Myosin", you can choose any protein you're interested in.
The first step to do is to enter to the SWISSPROT website at expasy:
1- Enter the site directly from here or go to google and write SWISSPROT, the first website at expasy is the SWISSPROT website, you'll see something like this:

2- Enter your protein name in the search box shown by the red arrow in the picture above.
* In this tutorial i'm searching for the protein "myosin" for example.
3- Click the GO button to start the search.
4- You will see the result page like bellow:


The information provided by this result is to huge and not accurate like related proteines, so we need to set a couple of things to get the results we need.
5- To do that you need to click on fields shown bellow:

You will see this:

From 1 shown by the arrow you can choose AND, NOT, OR
AND: means that you will add something like organism name for example.
NOT: means that you can eliminate searches that contain the word you'll write, like eliminate an organism.
OR: will searches for example for myosin OR actin if you want to.
From 2 we can choose our field like protein name, organism, gene name...etc
From 3 the term section, we can add the word we need to add it to search.
6- We will set AND and Protein name from the field dropdown menu.
7- In the term section we write Myosin to search only for Protein wich names are Myosin and exclude related proteines.
8- We click Add & Search, and we will see this:


We remark that the number of hits had dropped down and also this hits shows only proteines with protein name containing the word Myosin.
Let's say for example that you want only the protein in a specific organism like Homo Sapiens, then we will repeat the steps from 5 to 8 by clicking "field", choosing AND, Organism from the field dropdown menu and write Homo Sapiens in the Term field.
9- we click Add & Search and we'll get this:

As you can see that the numbet of hits has dropped from more than 200 in the first search to 11 here.
Because the protein Myosin have several chains, we will choose for example Myosin-Va.
10- By clicking the accession number shown bellow you will be taken to the information of this entry.

The informations about this protein are classed by category:
* Names and origins.
* Protein attributes.
* General anotations (protein function, subunits structure...etc).
* Refferences.
Our interest for now is the Sequences section, where we'll find the protein sequence.
We can see the sequence bellow:

To see the protein sequence without numbers within lines click on 1 shown by the red arrow
To do a blast search for similarities with other proteines choose Blast and click go like shown 2 by the red arrow.
Use SWISSPROT to search for a specific protein
(Video Lesson on Youtube)
(Video Lesson on Youtube)
Any comment or question, you're welcome.




0 comments:
Post a Comment